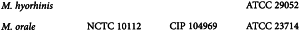Appendix XVI B (Vet) 3. Test for Absence of Mycoplasmas
Where the test for mycoplasmas is prescribed for a master cell bank, for a working cell bank, for a virus seed lot or for control cells, both the culture method and the indicator cell culture method are used. Where the test for mycoplasmas is prescribed for a virus harvest, for a bulk vaccine or for the final lot (batch), the culture method is used. The indicator cell culture method may also be used, where necessary, for screening of media.
Nucleic acid amplification techniques (NAT) may be used as an alternative to one or both of the other methods after suitable validation.
CULTURE METHOD
CHOICE OF CULTURE MEDIA
The test is carried out using a sufficient number of both solid and liquid media to ensure growth in the chosen incubation conditions of small numbers of mycoplasmas that may be present in the product to be examined. Liquid media must contain phenol red. The range of media chosen is shown to have satisfactory nutritive properties for at least the micro-organisms shown below. The nutritive properties of each new batch of medium are verified for the appropriate micro-organisms in the list. When testing for mycoplasmas in the product to be examined, at least 1 of the following species will be included as a positive control:
The test strains are field isolates having undergone a limited number of subcultures (not more than 15), and are stored frozen or freeze-dried. After cloning, the strains are identified as being of the required species by comparison with type cultures, for example:
Acholeplasma laidlawii BRP, Mycoplasma fermentans BRP, Mycoplasma hyorhinis BRP, Mycoplasma orale BRP and Mycoplasma synoviae BRP are suitable for use as low-passage reference strains.
INCUBATION CONDITIONS
Incubate liquid media in tightly stoppered containers at 35-38 °C. Incubate solid media in microaerophilic conditions (nitrogen containing 5-10 per cent of carbon dioxide and sufficient humidity to prevent desiccation of the agar surface) at 35-38 °C.
NUTRITIVE PROPERTIES
Carry out the test for nutritive properties for each new batch of medium Inoculate the chosen media with the appropriate test micro-organisms; use not more than 100 CFU per 60 mm diameter plate containing 9 mL of solid medium and per 100 mL container of liquid medium; use a separate plate and container for each species of micro-organism. Incubate the media and make subcultures from 0.2 mL of liquid medium to solid medium at the specified intervals (see below under Test for mycoplasmas in the product to be examined). The solid medium complies with the test if adequate growth is found for each test micro-organism (growth obtained does not differ by a factor greater than 5 from the value calculated with respect to the inoculum). The liquid medium complies with the test if growth on agar plates subcultured from the broth is found for at least 1 subculture for each test micro-organism.
INHIBITORY SUBSTANCES
The test for inhibitory substances is carried out once for a given product and is repeated whenever there is a change in production method that may affect the detection of mycoplasmas.
To demonstrate absence of inhibitory substances, carry out the test for nutritive properties in the presence and absence of the product to be examined. If growth of a test micro-organism occurs more than 1 subculture sooner in the absence of the product to be examined than in its presence, or if plates directly inoculated with the product to be examined have fewer than 1/5 of the number of colonies of those inoculated without the product to be examined, inhibitory substances are present and they must be neutralised or their effect otherwise countered, for example by passage in substrates not containing inhibitors or dilution in a larger volume of medium before the test. If dilution is used, larger medium volumes may be used or the inoculum volume may be divided among several 100 mL flasks. The effectiveness of the neutralisation or other process is checked by repeating the test for inhibitory substances after neutralisation.
TEST FOR MYCOPLASMAS IN THE PRODUCT TO BE EXAMINED
Inoculate 10 mL of the product to be examined per 100 mL of each liquid medium. If it has been found that a significant pH change occurs upon the addition of the product to be examined, the liquid medium is restored to its original pH value by the addition of a solution of either sodium hydroxide or hydrochloric acid. Inoculate 0.2 mL of the product to be examined on each plate of each solid medium. Incubate liquid media for 20-21 days. Incubate solid media for not less than 14 days, except those corresponding to the 20-21 day subculture, which are incubated for 7 days. At the same time incubate an uninoculated 100 mL portion of each liquid medium and agar plates, as a negative control. On days 2-4 after inoculation, subculture each liquid medium by inoculating 0.2 mL on at least 1 plate of each solid medium. Repeat the procedure between the 6th and 8th days, again between the 13th and 15th days and again between the 19th and 21st days of the test. Observe the liquid media every 2 or 3 days and if a colour change occurs, subculture. If a liquid medium shows bacterial or fungal contamination, the test is invalid. The test is valid if at least 1 plate per medium and per inoculation day can be read. Include in the test positive controls prepared by inoculation of not more than 100 CFU of at least 1 test micro-organism on agar medium or into broth medium. Where the test for mycoplasmas is carried out regularly and where possible, it is recommended to use the test micro-organisms in regular rotation. The test micro-organisms used are those listed under Choice of culture media.
interpretation of results
At the end of the prescribed incubation period, examine all inoculated solid media microscopically for the presence of mycoplasma colonies. The product complies with the test if growth of typical mycoplasma colonies has not occurred. The product does not comply with the test if growth of typical mycoplasma colonies has occurred on any of the solid media. The test is invalid if 1 or more of the positive controls do not show growth of mycoplasmas on at least 1 subculture plate. The test is invalid if 1 or more of the negative controls show growth of mycoplasmas. If suspect colonies are observed, a suitable validated method may be used to determine whether they are due to mycoplasmas.
The following section is published for information.
RECOMMENDED MEDIA FOR THE CULTURE METHOD
The following media are recommended. Other media may be used, provided that their ability to sustain the growth of mycoplasmas has been demonstrated on each batch in the presence and absence of the product to be examined.
Hayflick media (recommended for the general detection of mycoplasmas)
Liquid medium
Adjust to pH 7.8.
Solid medium
Prepare as described above replacing beef heart infusion broth by beef heart infusion agar containing 15 g/L of agar.
Frey media (recommended for the detection of m. Synoviae)
Liquid medium
Mix the solutions of β-nicotinamide adenine dinucleotide and cysteine hydrochloride and after 10 min add to the other ingredients. Adjust to pH 7.8.
Solid medium
Adjust to pH 7.8, sterilise by autoclaving then add:
FRIIS MEDIA (Recommended for the detection of non-avian mycoplasmas)
Liquid medium
Adjust to pH 7.40-7.45.
Solid medium
Mix well and sterilise by autoclaving. Cool to 100 °C. Add to 1740 mL of liquid medium as described above.
(1) Beef heart infusion broth
Sterilise by autoclaving.
(2) Essential vitamins
(3) Agar, purified
A highly refined agar for use in microbiology and immunology, prepared by an ion-exchange procedure that results in a product having superior purity, clarity and gel strength. It contains about:
(4) Hanks’ balanced salt solution (modified)
(5) Brain heart infusion
(6) PPLO broth
INDICATOR CELL CULTURE METHOD
Cell cultures are stained with a fluorescent dye that binds to DNA. Mycoplasmas are detected by their characteristic particulate or filamentous pattern of fluorescence on the cell surface and, if contamination is heavy, in surrounding areas. Mitochondria in the cytoplasm may be stained but are readily distinguished from mycoplasmas.
If for viral suspensions the interpretation of results is affected by marked cytopathic effects, the virus may be neutralised using a specific antiserum that has no inhibitory effects on mycoplasmas or a cell culture substrate that does not allow growth of the virus may be used. To demonstrate the absence of inhibitory effects of serum, carry out the positive control tests in the presence and absence of the antiserum.
VERIFICATION OF THE SUBSTRATE
Use Vero cells or another cell culture (for example, the production cell line) that is equivalent in effectiveness for detecting mycoplasmas. Test the effectiveness of the cells to be used by applying the procedure shown below and inoculating not more than 100 CFU or CFU-like micro-organisms of suitable reference strains of M. hyorhinis and M. orale. The following strains have been found to be suitable:
The cells are suitable if both reference strains are detected.
The indicator cells must be subcultured without an antibiotic before use in the test.
TEST METHOD
INTERPRETATION OF RESULTS
The product to be examined complies with the test if fluorescence typical of mycoplasmas is not present. The test is invalid if the positive controls do not show fluorescence typical of mycoplasmas. The test is invalid if the negative controls show fluorescence typical of mycoplasmas.
nucleic acid amplification techniques (nat)
NAT (2.6.21) may be used for detection of mycoplasmas by amplification of nucleic acids extracted from a test sample with specific primers that reveal the presence of the target nucleic acid. NAT indicate the presence of a particular nucleic acid sequence and not necessarily the presence of viable mycoplasmas. A number of different techniques are available. This general chapter does not prescribe a particular method for the test. The procedure applied must be validated as described, taking account of the guidelines presented at the end of this section. Where a commercial kit is used, certain elements of the validation may be carried out by the manufacturer and information provided to the user but it must be remembered that full information on the primers may not be available and that production of the kit may be modified or discontinued.
NAT are applied where prescribed in a monograph. They may also be used instead of the culture method and the indicator cell culture method after suitable validation.
Direct NAT Can be applied in the presence of cytotoxic material and where a rapid method is needed.
Cell-culture enrichment followed by NAT The test sample and a suitable cell substrate (as described under the indicator cell-culture method) are cultured together for a suitable period; the nucleic acids are then extracted from cells and supernatant and used for detection by NAT.
Validation
Reference standards are required at various stages during validation and for use as controls during routine application of the test. The reference standards may be mycoplasmas or nucleic acids.
For validation of the limit of detection, the following species represent an optimal selection in terms of the frequency of occurrence as contaminants and phylogenetic relationships:
Demonstration of specificity requires the use of a suitable range of bacterial species other than mycoplasmas. Bacterial genera with close phylogenetic relation to mycoplasmas are most appropriate for this validation; these include Clostridium, Lactobacillus and Streptococcus.
Comparability studies for use of NAT as an alternative method For each mycoplasma test species:
or an equivalent limit of detection in terms of the number of copies of mycoplasma nucleic acid in the test sample (using suitable reference standards of mycoplasma nucleic acid).
Controls
Internal controls
Internal controls are necessary for routine verification of absence of inhibition. The internal control may contain the primer binding-site, or some other suitable sequence may be used. It is preferably added to the test material before isolating the nucleic acid and therefore acts as an overall control (extraction, reverse transcription, amplification, detection).
External controls
The external positive control contains a defined number of target-sequence copies or CFUs from 1 or more suitable species of mycoplasma chosen from those used during validation of the test conditions. 1 of the positive controls is set close to the positive cut-off point to demonstrate that the expected sensitivity is achieved. The external negative control contains no target sequence but does not necessarily represent the same matrix as the test article.
Interpretation of results
The primers used may also amplify non-mycoplasmal bacterial nucleic acid, leading to false positive results. Procedures are established at the time of validation for dealing with confirmation of positive results, where necessary.
The following section is published for information.
Validation of nucleic acid amplification techniques (NAT) for the detection of mycoplasmas: guidelines
1 SCOPE
Nucleic acid amplification techniques (NAT) are either qualitative or quantitative tests for the presence of nucleic acid. For the detection of mycoplasma contamination of various samples such as vaccines and cell substrates, qualitative tests are adequate and may be considered to be limit tests.
These guidelines describe methods to validate qualitative nucleic acid amplification analytical procedures for assessing mycoplasma contamination. They may also be applicable for real-time NAT used as limit tests for the control of contaminants.
The 2 characteristics regarded as the most important for validation of the analytical procedure are the specificity and the detection limit. In addition, the robustness of the analytical procedure should be evaluated.
For the purpose of this document, an analytical procedure is defined as the complete procedure from extraction of nucleic acid to detection of the amplified products.
Where commercial kits are used for part or all of the analytical procedure, documented validation points already covered by the kit manufacturer can replace validation by the user. Nevertheless, the performance of the kit with respect to its intended use has to be demonstrated by the user (e.g. detection limit, robustness, cross-detection of other classes of bacteria).
NAT may be used as:
These guidelines will thus separate these 2 objectives by presenting first a guideline for the validation of the NAT themselves, and second, a guideline for a comparability study between NAT and official methods.
2 Guideline for mycoplasma nat validation
3 parameters should be evaluated: specificity, detection limit and robustness.
2-1 Specificity
Specificity is the ability to unequivocally assess target nucleic acid in the presence of components that may be expected to be present.
The specificity of NAT is dependent on the choice of primers, the choice of probe (for analysis of the final product) and the stringency of the test conditions (for both the amplification and detection steps).
The ability of the NAT to detect a large panel of mycoplasma species will depend on the choice of primers, probes and method parameters. This ability should be demonstrated using characterised reference panels (e.g. reference strains provided by the EDQM). Since NAT systems are usually based on a mix of primers, the theoretical analysis of primers and probes by comparison with databases is not recommended, because interpretation of the results may be quite complex and may not reflect the experimental results.
Moreover, as it is likely that the primers will detect other bacterial species, the potential cross-detection should be documented in the validation study. Bacterial genera such as gram-positive bacteria with close phylogenetic relation to mycoplasmas are most appropriate for this validation; these include Clostridium, Lactobacillus and Streptococcus. However, this is not an exhaustive list and species to be tested will depend on the theoretical ability (based on primers/probes sequences) of the NAT system to detect such other species.
Based on the results from this validation of the specificity, if a gap in the specificity of the method is identified (such as detection of non-mycoplasmal bacterial nucleic acid), an appropriate strategy must be proposed in the validation study to allow interpretation of positive results on a routine basis. For example, a second test may be performed using an alternative method without this specificity gap or using an official method.
2-2 Detection limit
The detection limit of an individual analytical procedure is the lowest amount of target nucleic acid in a sample that can be detected but not necessarily quantitated as an exact value.
For establishment of the detection limit, a positive cut-off point should be determined for the nucleic acid amplification analytical procedure. The positive cut-off point (as defined in general chapter 2.6.21) is the minimum number of target sequence copies per volume of sample that can be detected in 95 per cent of test runs. This positive cut-off point is influenced by the distribution of mycoplasmal genomes in the individual samples being tested and by factors such as enzyme efficiency, and can result in different 95 per cent cut-off values for individual analytical test runs.
To determine the positive cut-off point, a dilution series of characterised and calibrated (either in CFUs or nucleic acid copies) in-house working strains or EDQM standards should be tested on different days to examine variation between test runs.
For validation of the limit of detection, the following species represent an optimal selection in terms of the frequency of occurrence as contaminants and phylogenetic relationships:
For each strain, at least 3 independent 10-fold dilution series should be tested, with a sufficient number of replicates at each dilution to give a total number of 24 test results for each dilution, to enable a statistical analysis of the results.
For example, a laboratory may test 3 dilution series on different days with 8 replicates for each dilution, 4 dilution series on different days with 6 replicates for each dilution, or 6 dilution series on different days with 4 replicates for each dilution. In order to keep the number of dilutions at a manageable level, a preliminary test should be performed to obtain a preliminary value for the positive cut-off point (i.e. the highest dilution giving a positive signal). The range of dilutions can then be chosen around the predetermined preliminary cut-off point. The concentration of mycoplasmas (CFUs or copies) that can be detected in 95 per cent of test runs can then be calculated using an appropriate statistical evaluation.
These results may also serve to evaluate the variability of the analytical procedure.
2-3 Robustness
The robustness of an analytical procedure is a measure of its capacity to remain unaffected by small but deliberate variations in method parameters, and provides an indication of its reliability during normal usage.
The evaluation of robustness should be considered during the development phase. It should show the reliability of the analytical procedure with respect to deliberate variations in method parameters. For NAT, small variations in the method parameters can be crucial. However, the robustness of the method can be demonstrated during its development when small variations in the concentrations of reagents (e.g. MgCl2, primers or deoxyribonucleotides) are tested. Modifications of extraction kits or extraction procedures as well as different thermal cycler types may also be evaluated.
Finally, robustness of the method can be evaluated through collaborative studies.
3 Guideline for comparability study
NAT may be used instead of official methods (indicator cell culture method and/or culture method). In this case a comparability study should be carried out. This comparability study should include mainly a comparison of the respective detection limits of the alternative method and official methods. However, specificity (mycoplasma panel detected, putative false positive results) should also be considered.
For the detection limit, acceptability criteria are defined as follows:
For both cases, suitable standards calibrated for the number of nucleic acid copies and the number of CFUs may be used for establishing that these acceptability criteria are reached. The relation between CFUs and nucleic acid copies for the reference preparations should be previously established to compare the performance of the alternative NAT method with the performance of the official methods.
1 of the following 2 strategies can be used to perform this comparability study:
Comparability study reports should describe all the validation elements described in section 2 (specificity, limit of detection and variability, as well as robustness) in order to assess all the advantages and/or disadvantages of the alternative NAT method compared to official methods.