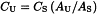Appendix VIII P. Total Protein
Many of the assay methods described in this chapter can be performed using kits from commercial sources.
METHOD 1
Protein in solution absorbs ultraviolet light at a wavelength of 280 nm, due to the presence of aromatic amino acids, mainly tyrosine and tryptophan, in the protein structure. This property can be used for assay purposes. If the buffer used to dissolve the protein has a high absorbance relative to that of water, an interfering substance is present. This interference may be obviated by using the buffer as compensation liquid but if the interfering substance produces a high absorbance, the results may nevertheless be compromised. At low concentrations, protein adsorbed onto the cell may significantly reduce the content in solution. This can be prevented by preparing samples at higher concentration or by using a non-ionic detergent in the preparation.
Test solution Dissolve a suitable quantity of the substance to be examined in the prescribed buffer to obtain a solution having a protein concentration between 0.2 mg/mL and 2 mg/mL.
Reference solution Prepare a solution of a suitable reference substance for the protein to be determined, in the same buffer and at the same protein concentration as the test solution.
Procedure
Keep the test solution, the reference solution and the compensation liquid at the same temperature during the performance of this test. Determine the absorbances (2.2.25) of the test solution and the reference solution in quartz cells at 280 nm, using the prescribed buffer as the compensation liquid. The response must be linear in the range of protein concentrations to be assayed to obtain accurate results.
Light scattering
The accuracy of the determination of protein can be diminished by the scattering of light by the test sample. If the proteins in solution exist as particles comparable in size to the wavelength of the measuring light (250 nm to 300 nm), scattering of the light beam results in an apparent increase in absorbance of the test sample. To calculate the absorbance at 280 nm due to light scattering, determine the absorbances of the test solution at wavelengths of 320 nm, 325 nm, 330 nm, 335 nm, 340 nm, 345 nm and 350 nm. Plot the logarithm of the observed absorbance against the logarithm of the wavelength and determine the standard curve best fitting the plotted points by linear regression. Extrapolate the curve to determine the logarithm of the absorbance at 280 nm. The antilogarithm of this value is the absorbance attributed to light scattering. Correct the observed values by subtracting the absorbance attributed to light scattering from the total absorbance at 280 nm to obtain the absorbance value of the protein in solution. Filtration with a 0.2 µm filter that does not adsorb protein or clarification by centrifugation may be performed to reduce the effect of light scattering, especially if the solution is noticeably turbid.
Calculations
Use corrected values for the calculations. Calculate the concentration of protein in the test solution (CU) from the following equation:
where CS is the concentration of protein in the reference solution and AU and AS are the corrected absorbances of the test solution and the reference solution, respectively.
METHOD 2
This method (commonly referred to as the Lowry assay) is based on the reduction by protein of the phosphomolybdotungstic mixed acid chromogen in the phosphomolybdotungstic reagent, which results in an absorbance maximum at 750 nm. The phosphomolybdotungstic reagent reacts primarily with tyrosine residues in the protein. Colour development reaches a maximum in 20 min to 30 min at room temperature, after which there is a gradual loss of colour. Because the method is sensitive to interfering substances, a procedure for precipitation of the protein from the test sample may be used. Most interfering substances cause a lower colour yield; however, some detergents cause a slight increase in colour. A high salt concentration may cause a precipitate to form. Because different protein species may give different colour response intensities, the reference substance and test protein must be the same. Where separation of interfering substances from the protein in the test sample is necessary, proceed as directed below for interfering substances prior to preparation of the test solution. The effect of interfering substances may be minimised by dilution, provided the concentration of the test protein remains sufficient for accurate measurement.
Use distilled water R to prepare all buffers and reagents used for this method.
Test solution Dissolve a suitable quantity of the substance to be examined in the prescribed buffer to obtain a solution having a concentration within the range of the standard curve. A suitable buffer will produce a solution of pH 10.0 to 10.5.
Reference solutions Dissolve the reference substance for the protein to be determined in the prescribed buffer. Dilute portions of this solution with the same buffer to obtain not fewer than five reference solutions having protein concentrations evenly spaced over a suitable range situated between 5 µg/mL and 100 µg/mL.
Blank Use the buffer used to prepare the test solution and the reference solutions.
Copper sulfate reagent Dissolve 100 mg of copper sulfate pentahydrate R and 0.2 g of sodium tartrate R in distilled water R and dilute to 50 mL with the same solvent. Dissolve 10 g of anhydrous sodium carbonate R in distilled water R and dilute to 50 mL with the same solvent. Slowly pour the sodium carbonate solution into the copper sulfate solution with mixing. Use within 24 h.
Alkaline copper reagent Mix 1 volume of copper sulfate reagent, 2 volumes of a 50 g/L solution of sodium dodecyl sulfate R and 1 volume of a 32 g/L solution of sodium hydroxide R. Store at room temperature and use within 2 weeks.
Diluted phosphomolybdotungstic reagent Mix 5 mL of phosphomolybdotungstic reagent R with 55 mL of distilled water R. Store in an amber bottle, at room temperature.
Procedure
To 1.0 mL of each reference solution, of the test solution and of the blank, add 1.0 mL of alkaline copper reagent and mix. Allow to stand for 10 min. Add 0.5 mL of the diluted phosphomolybdotungstic reagent, mix and allow to stand at room temperature for 30 min. Determine the absorbances (2.2.25) of the solutions at 750 nm, using the solution from the blank as compensation liquid.
Calculations
The relationship of absorbance to protein concentration is non-linear; however, if the range of concentrations used to prepare the standard curve is sufficiently small, the latter will approach linearity. Plot the absorbances of the reference solutions against the protein concentrations and use linear regression to establish the standard curve. From the standard curve and the absorbance of the test solution, determine the concentration of protein in the test solution.
Interfering substances
In the following procedure, deoxycholate-trichloroacetic acid is added to a test sample to remove interfering substances by precipitation of proteins before determination; this technique can also be used to concentrate proteins from a dilute solution.
Add 0.1 mL of a 1.5 g/L solution of sodium deoxycholate R to 1 mL of a solution of the substance to be examined. Mix using a vortex mixer and allow to stand at room temperature for 10 min. Add 0.1 mL of a 720 g/L solution of trichloroacetic acid R and mix using a vortex mixer. Centrifuge at 3000 g for 30 min, decant the liquid and remove any residual liquid with a pipette. Redissolve the protein pellet in 1 mL of alkaline copper reagent.
METHOD 3
This method (commonly referred to as the Bradford assay) is based on the absorption shift from 470 nm to 595 nm observed when the acid blue 90 dye binds to protein. The acid blue 90 dye binds most readily to arginine and lysine residues in the protein which can lead to variation in the response of the assay to different proteins. The protein used as reference substance must therefore be the same as the protein to be determined. There are relatively few interfering substances, but it is preferable to avoid detergents and ampholytes in the test sample. Highly alkaline samples may interfere with the acidic reagent.
Use distilled water R to prepare all buffers and reagents used for this method.
Test solution Dissolve a suitable quantity of the substance to be examined in the prescribed buffer to obtain a solution having a concentration within the range of the standard curve.
Reference solutions Dissolve the reference substance for the protein to be determined in the prescribed buffer. Dilute portions of this solution with the same buffer to obtain not fewer than five reference solutions having protein concentrations evenly spaced over a suitable range situated between 0.1 mg/mL and 1 mg/mL.
Blank Use the buffer used to prepare the test solution and the reference solutions.
Acid blue 90 reagent Dissolve 0.10 g of acid blue 90 R in 50 mL of alcohol R. Add 100 mL of phosphoric acid R, dilute to 1000 mL with distilled water R and mix. Filter the solution and store in an amber bottle at room temperature. Slow precipitation of the dye occurs during storage. Filter the reagent before using.
Procedure
Add 5 mL of acid blue 90 reagent to 0.100 mL of each reference solution, of the test solution and of the blank. Mix by inversion. Avoid foaming, which will lead to poor reproducibility. Determine the absorbances (2.2.25) of the standard solutions and of the test solution at 595 nm, using the blank as compensation liquid. Do not use quartz (silica) spectrophotometer cells because the dye binds to this material.
Calculations
The relationship of absorbance to protein concentration is non-linear; however, if the range of concentrations used to prepare the standard curve is sufficiently small, the latter will approach linearity. Plot the absorbances of the reference solutions against protein concentrations and use linear regression to establish the standard curve. From the standard curve and the absorbance of the test solution, determine the concentration of protein in the test solution.
METHOD 4
This method (commonly referred to as the bicinchoninic acid or BCA assay) is based on reduction of the cupric (Cu2+) ion to cuprous (Cu1+) ion by protein. The bicinchoninic acid reagent is used to detect the cuprous ion. Few substances interfere with the reaction. When interfering substances are present their effect may be minimised by dilution, provided that the concentration of the protein to be determined remains sufficient for accurate measurement. Alternatively, the protein precipitation procedure given in Method 2 may be used to remove interfering substances. Because different protein species may give different colour response intensities, the reference protein and protein to be determined must be the same.
Use distilled water R to prepare all buffers and reagents used for this method.
Test solution Dissolve a suitable quantity of the substance to be examined in the prescribed buffer to obtain a solution having a concentration within the range of the concentrations of the reference solutions.
Reference solutions Dissolve the reference substance for the protein to be determined in the prescribed buffer. Dilute portions of this solution with the same buffer to obtain not fewer than five reference solutions having protein concentrations evenly spaced over a suitable range situated between 10 µg/mL and 1200 µg/mL.
Blank Use the buffer used to prepare the test solution and the reference solutions.
BCA reagent Dissolve 10 g of disodium bicinchoninate R, 20 g of sodium carbonate monohydrate R, 1.6 g of sodium tartrate R, 4 g of sodium hydroxide R, and 9.5 g of sodium hydrogen carbonate R in distilled water R. Adjust, if necessary, to pH 11.25 with a solution of sodium hydroxide R or a solution of sodium hydrogen carbonate R. Dilute to 1000 mL with distilled water R and mix.
Copper-BCA reagent Mix 1 mL of a 40 g/L solution of copper sulfate pentahydrate R and 50 mL of BCA reagent.
Procedure
Mix 0.1 mL of each reference solution, of the test solution and of the blank with 2 mL of the copper-BCA reagent. Incubate the solutions at 37 °C for 30 min, note the time and allow the mixtures to cool to room temperature. Within 60 min of the end of incubation, determine the absorbances (2.2.25) of the reference solutions and of the test solution in quartz cells at 562 nm, using the blank as compensation liquid. After the solutions have cooled to room temperature, the colour intensity continues to increase gradually.
Calculations
The relationship of absorbance to protein concentration is non-linear; however, if the range of concentrations used to prepare the standard curve is sufficiently small, the latter will approach linearity. Plot the absorbances of the reference solutions against protein concentrations and use linear regression to establish the standard curve. From the standard curve and the absorbance of the test solution, determine the concentration of protein in the test solution.
METHOD 5
This method (commonly referred to as the biuret assay) is based on the interaction of cupric (Cu2+) ion with protein in alkaline solution and resultant development of absorbance at 545 nm. This test shows minimal difference between equivalent IgG and albumin samples. Addition of the sodium hydroxide and the biuret reagent as a combined reagent, insufficient mixing after the addition of the sodium hydroxide, or an extended time between the addition of the sodium hydroxide solution and the addition of the biuret reagent will give IgG samples a higher response than albumin samples. The trichloroacetic acid method used to minimise the effects of interfering substances also can be used to determine the protein content in test samples at concentrations below 500 µg/mL.
Use distilled water R to prepare all buffers and reagents used for this method.
Test solution Dissolve a suitable quantity of the substance to be examined in a 9 g/L solution of sodium chloride R to obtain a solution having a concentration within the range of the concentrations of the reference solutions.
Reference solutions Dissolve the reference substance for the protein to be determined in a 9 g/L solution of sodium chloride R. Dilute portions of this solution with a 9 g/L solution of sodium chloride R to obtain not fewer than three reference solutions having protein concentrations evenly spaced over a suitable range situated between 0.5 mg/mL and 10 mg/mL.
Blank Use a 9 g/L solution of sodium chloride R.
Biuret reagent Dissolve 3.46 g of copper sulfate pentahydrate R in 10 mL of hot distilled water R, and allow to cool (Solution A). Dissolve 34.6 g of sodium citrate R and 20.0 g of anhydrous sodium carbonate R in 80 mL of hot distilled water R, and allow to cool (Solution B). Mix solutions A and B and dilute to 200 mL with distilled water R. Use within 6 months. Do not use the reagent if it develops turbidity or contains any precipitate.
Procedure
To one volume of the test solution add an equal volume of a 60 g/L solution of sodium hydroxide R and mix. Immediately add biuret reagent equivalent to 0.4 volumes of the test solution and mix rapidly. Allow to stand at a temperature between 15 °C and 25 °C for not less than 15 min. Within 90 min of addition of the biuret reagent, determine the absorbances (2.2.25) of the reference solutions and of the test solution at the maximum at 545 nm, using the blank as compensation liquid. Any solution that develops turbidity or a precipitate is not acceptable for calculation of protein concentration.
Calculations
The relationship of absorbance to protein concentration is approximately linear within the indicated range of protein concentrations for the reference solutions. Plot the absorbances of the reference solutions against protein concentrations and use linear regression to establish the standard curve. Calculate the correlation coefficient for the standard curve. A suitable system is one that yields a line having a correlation coefficient not less than 0.99. From the standard curve and the absorbance of the test solution, determine the concentration of protein in the test solution.
Interfering substances
To minimise the effect of interfering substances, the protein can be precipitated from the test sample as follows: add 0.1 volumes of a 500 g/L solution of trichloroacetic acid R to 1 volume of a solution of the test sample, withdraw the supernatant layer and dissolve the precipitate in a small volume of 0.5 M sodium hydroxide. Use the solution obtained to prepare the test solution.
METHOD 6
This fluorimetric method is based on the derivatisation of the protein with o-phthalaldehyde, which reacts with the primary amines of the protein (N-terminal amino acid and the ε-amino group of lysine residues). The sensitivity of the assay can be increased by hydrolysing the protein before adding o-phthalaldehyde. Hydrolysis makes the α-amino group of the constituent amino acids available for reaction with the phthalaldehyde reagent. The method requires very small quantities of the protein. Primary amines, such as tris(hydroxymethyl)aminomethane and amino acid buffers, react with phthalaldehyde and must be avoided or removed. Ammonia at high concentrations reacts with phthalaldehyde. The fluorescence obtained when amine reacts with phthalaldehyde can be unstable. The use of automated procedures to standardise this procedure may improve the accuracy and precision of the test.
Use distilled water R to prepare all buffers and reagents used for this method.
Test solution Dissolve a suitable quantity of the substance to be examined in a 9 g/L solution of sodium chloride R to obtain a solution having a concentration within the range of the concentrations of the reference solutions. Adjust the solution to pH 8 to 10.5 before addition of the phthalaldehyde reagent.
Reference solutions Dissolve the reference substance for the protein to be determined in a 9 g/L solution of sodium chloride R. Dilute portions of this solution with a 9 g/L solution of sodium chloride R to obtain not fewer than five reference solutions having protein concentrations evenly spaced over a suitable range situated between 10 µg/mL and 200 µg/mL. Adjust the solutions to pH 8 to 10.5 before addition of the phthalaldehyde reagent.
Blank solution Use a 9 g/L solution of sodium chloride R.
Borate buffer solution Dissolve 61.83 g of boric acid R in distilled water R and adjust to pH 10.4 with a solution of potassium hydroxide R. Dilute to 1000 mL with distilled water R and mix.
Phthalaldehyde stock solution Dissolve 1.20 g of phthalaldehyde R in 1.5 mL of methanol R, add 100 mL of borate buffer solution and mix. Add 0.6 mL of a 300 g/L solution of macrogol 23 lauryl ether R and mix. Store at room temperature and use within 3 weeks.
Phthalaldehyde reagent To 5 mL of phthalaldehyde stock solution add 15 µL of 2-mercaptoethanol R. Prepare at least 30 min before use. Use within 24 h.
Procedure
Mix 10 µL of the test solution and of each of the reference solutions with 0.1 mL of phthalaldehyde reagent and allow to stand at room temperature for 15 min. Add 3 mL of 0.5 M sodium hydroxide and mix. Determine the fluorescent intensities (2.2.21) of solutions from the reference solutions and from the test solution at an excitation wavelength of 340 nm and an emission wavelength between 440 and 455 nm. Measure the fluorescent intensity of a given sample only once, since irradiation decreases the fluorescence intensity.
Calculations
The relationship of fluorescence to protein concentration is linear. Plot the fluorescent intensities of the reference solutions against protein concentrations and use linear regression to establish the standard curve. From the standard curve and the fluorescent intensity of the test solution, determine the concentration of protein in the test solution.
METHOD 7
This method is based on nitrogen analysis as a means of protein determination. Interference caused by the presence of other nitrogen-containing substances in the test sample can affect the determination of protein by this method. Nitrogen analysis techniques destroy the test sample during the analysis but are not limited to protein presentation in an aqueous environment.
Procedure A
Proceed as prescribed for the determination of nitrogen by sulfuric acid digestion (2.5.9) or use commercial instrumentation for Kjeldahl nitrogen assay.
Procedure B
Commercial instrumentation is available for nitrogen analysis. Most nitrogen analysis instruments use pyrolysis (i.e. combustion of the sample in oxygen at temperatures approaching 1000 °C), which produces nitric oxide (NO) and other oxides of nitrogen (NOx) from the nitrogen present in the substance to be examined. Some instruments convert the nitric oxides to nitrogen gas, which is quantified using a thermal-conductivity detector. Other instruments mix nitric oxide (NO) with ozone (O3) to produce excited nitrogen dioxide (NO2*), which emits light when it decays and can be quantified with a chemiluminescence detector. A protein reference material that is relatively pure and is similar in composition to the test proteins is used to optimise the injection and pyrolysis parameters and to evaluate consistency in the analysis.
Calculations
The protein concentration is calculated by dividing the nitrogen content of the sample by the known nitrogen content of the protein. The known nitrogen content of the protein can be determined from the chemical composition of the protein or by comparison with a suitable reference substance.