SC VII D. DNA Barcoding as a tool for Botanical Identification of Herbal Drugs
This Supplementary Chapter concerns the use of DNA barcoding as a tool for botanical identification of herbal drugs. For the purposes of identification, DNA barcoding has been widely accepted as sufficiently robust and is considered an appropriate method for molecular identification of herbal materials.
Introduction
In generating the reference barcode sequences published for a herbal drug, several barcode regions are examined including the nuclear Internal Transcribed Spacer (nrITS) and several plastid regions; trnH-psbA, rbcL, trnL-F and matK. The most informative region is identified for each herbal drug, based on the specificity of the DNA sequence to the named species. The species specific sequence for that region is published, highlighting those bases that are essential for identification. Further barcode regions are examined when no species specific sequences occur in the regions named above.
When a DNA-based identification technique is specified in a monograph for a herbal drug, the identified barcode region and its species specific sequence (reference sequence) will be published as part of the monograph. Where the barcode analysis for a herbal drug has been completed by the BP, but the DNA-based identification technique is not specified as an identification method in the monograph, the barcode region and reference sequence will form part of this Supplementary Chapter.
The usual method of obtaining a barcode sequence from plant material is by DNA extraction, Polymerase Chain Reaction (PCR) and Sanger sequencing. Appendix XI V ‘Deoxyribonucleic Acid (DNA) Based Identification Techniques for Herbal Drugs’ provides further details. This Appendix encompasses the theory of DNA-based techniques, the infrastructure required, controls and general methods. When a reference sequence is published in a herbal drug monograph or in this Supplementary Chapter the chosen barcode region and any deviations from the published Appendix XI V method for that barcode region will be given either in the herbal drug monograph or in this Supplementary Chapter, as applicable.
Sanger Sequencing: Application and Limitations
Sanger sequencing is the most widely used and easily accessible form of DNA sequencing. However, there are limitations to this technique when applied to herbal drugs, including the inability to detect adulterants, lack of information on purity, potential problems of application to degraded DNA samples, and higher cost, time and technical skill requirements when compared with other DNA-based techniques. To address these limitations, the feasibility of the use of alternative techniques is being investigated and will be applied when appropriate. For example, where adulteration is known to be an issue, consideration will be given to designing a technique that identifies and quantifies the target species and the adulterant species simultaneously. This Supplementary Chapter will be updated to reflect other DNA-based methods as and when they are applied within the BP.
Reference Material – British Pharmacopoeia Nucleic Acid Reference Material (BPNARM)
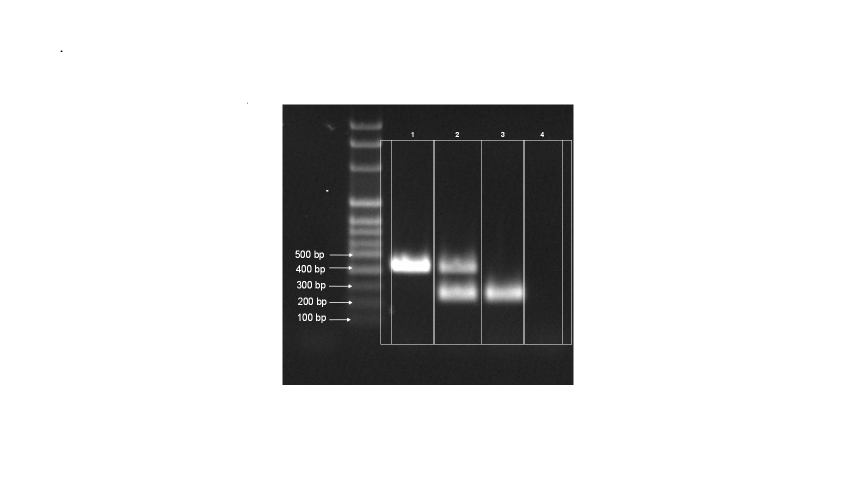
To confirm the suitability of the DNA extraction, the PCR chemistry and working practices, appropriate British Pharmacopoeia Nucleic Acid Reference Materials (BPNARM) will be available to end users. Reference electrophoresis results (Figure 1) will be produced in the leaflets for BPNARMs showing the pattern of banding that satisfies the criteria for acceptance.
Table 1
Reference Electrophoresis Results
Lane 1 | Amplification product from herbal drug |
Lane 2 | Amplification product showing both the herbal drug and the BPNARM band. This is the DNA extraction control |
Lane 3 | Amplification product of the BPNARM alone, the PCR positive control |
Lane 4 | Negative control with no amplification product visible |
Table 2
Criteria for Acceptance
Lane 1 | Visible band from the plant DNA |
Lane 2 | 245 bp band from the BPNARM. The band from the plant DNA may or may not be present |
Lane 3 | 245 bp band from the BPNARM |
Lane 4 | No band |
The suitability of the DNA extraction process used is shown by the results in Lanes 1 to 3 by the banding pattern present. More specifically the BPNARM band is shown in Lanes 2 and 3. As this material is provided as a known DNA sample that amplifies with the method given, an absence of the 245 bp band in both Lanes 2 and 3 shows that the process has not been completed satisfactorily. It is possible for compounds present in the herbal drug to co-purify with the DNA sample, inhibiting the PCR so that no band is formed. Should this occur the subsequent banding pattern would show no bands in Lane 2, due to inhibition, but a 245 bp band in Lane 3, because no plant material is present and therefore no inhibitory compounds. Without the use of the BPNARM it is difficult to discern whether the DNA extraction or the presence of inhibitory compounds has caused the failure of the PCR. Another possibility is that the DNA in the herbal drug may be too degraded to amplify; this would result in the BPNARM bands being formed, but no bands from the plant DNA (Lanes 1 and 2 in Figure 1). The suitability of the PCR chemistry and instrumentation used is verified by the production of the 245 bp BPNARM band in Lane 3. The reaction that produces the band in Lane 3 is the PCR positive control. If a band is not produced from this reaction then there is a problem with either: the components used to make up the reaction, the thermal cycler or the program entered into it, or a user error in the preparation of the reaction mixture.
The suitability of the working practices used to produce the PCRs and the agarose gel electrophoresis system by which to view the amplification products is verified by the presence of a band from the positive control reaction and also by the absence of any other unexplained banding. Lane 4 on Figure 1 is the PCR ‘negative control’ for which the acceptance criterion is the absence of a band. Should a band be formed in Lane 4, this is likely to be due to contamination of the PCR with extraneous DNA. Similarly, if an unexplained band is found in any of the lanes, this may show contamination of the PCR and/or the DNA extraction, or the adulteration of the herbal drug from which the DNA was extracted.
For the Ocimum tenuiflorum barcoding method specified in Appendix XI V a reference material trnH-psbA BPNARM has been developed and is listed in the BP Reference Standards Catalogue. This is available through the BP Website (http://www.pharmacopoeia.com). A BPNARM is not tied to one species, but to the chosen DNA barcode region. Therefore, trnH-psbA BPNARM will be applicable to all DNA-based identification techniques based on sequencing the trnH-psbA plastid region.
Reference Sequences
When reference DNA sequences are provided, either as one of the identification methods in a monograph or in this Supplementary Chapter, they are published with the key bases for identification indicated by lower case text. In some reference sequences the degenerate DNA code is used, this gives information about the permitted bases at any one position when variation is known to occur. For example, in sequences where variation is seen between a Cytosine (C) and a Thymine (T) at a base position, the degenerate code Y is shown. This means that either a Cytosine (C) or a Thymine (T) would be permitted at the base position, but rules out a Guanine (G) or an Adenine (A) base. The full degenerate code is shown in Table 3.
Table 3
Degenerate DNA Code
Code | Meaning |
K | G or T |
M | A or C |
R | A or G |
Y | C or T |
S | C or G |
W | A or T |
B | C or G or T |
V | A or C or G |
H | A or C or T |
D | A or G or T |
The matching of DNA sequences produced from samples against a published BP Reference Sequence can be achieved by the use of DNA sequence alignment software. Many applications are available for this including internet based, free access portals. Query sequences should be aligned with the relevant BP Reference Sequence and the results are given in both percentage similarity of sequences and full images of the entire alignment. It is possible to upload query sequences from multiple samples to be aligned with a BP Reference Sequence simultaneously. Careful attention must be paid to the key bases for identification as these must match exactly and be checked manually.
The BP Reference Sequences are provided along with any additional information considered helpful to the analyst in the analysis of samples using an Appendix XI V method. This will include adaptations to optimise DNA extraction and purification methods and details of sequencing primers. Sequencing is often carried out using the PCR amplification primers, but it can be useful to use different primers at this stage depending on the DNA sequence.
trnH-psbA Barcode Region Reference Sequences
Phellodendron Amurense Bark
Phellodendron amurense Ruprecht
Due to the presence of polysaccharides, DNA extraction is optimal using less starting material, for example 2 mg rather than 20 mg.
Sequencing Primers
Forward 5ʹ CCATGAAGATCGAAGGGCAC 3ʹ
Reverse 5ʹ GGGGGTCGGTATTAATCCGTT 3ʹ
Reference Sequence:
Phellodendron chinense Schneid, a closely related species, is known to occur as a substitute for or as an adulterant in Phellodendron amurense. The barcode reference sequence for Phellodendron chinense is presented below.
Reference Sequence:
Phellodendron Chinense Bark
Phellodendron chinense Schneid
Due to the presence of polysaccharides, DNA extraction is optimal using less starting material, for example 2 mg rather than 20 mg.
Sequencing Primers
92 F 5ʹ CCATGAAGATCGAAGGGCAC 3ʹ
358 R 5ʹ GGGGGTCGGTATTAATCCGTT 3ʹ
Reference Sequence:
Phellodendron amurense Ruprecht, a closely related species, is known to occur as a substitute for or as an adulterant in Phellodendron chinense. The barcode reference sequence for Phellodendron amurense is presented below.
Reference Sequence:
GLOSSARY
Table 4
Term | Definition | |
Amplicon | The DNA product of a PCR. | |
Amplification | The copying of DNA during a PCR. | |
Base call | The identification of a DNA base by sequencing software. | |
Base pair (bp) | The complementary pairing of two nucleotides, A&T or G&C, which forms the unit of measurement for the length of a DNA molecule. | |
BPNARM | British Pharmacopoeia Nucleic Acid Reference Material. | |
Consensus sequence | The product of the combining of several individual DNA sequencing reads, providing a consensus of the correct sequence. | |
Contig | A set of overlapping DNA sequencing reads from one sample which can be used to produce a consensus sequence. | |
Deoxynucleotide (dNTP) | The monomer or individual unit of DNA; Adenine (A), Cytosine (C), Guanine (G) and Thymine (T). | |
di-deoxynucleotide (ddNTP) | A modified form of the DNA monomer without an -OH group present on the 3ʹ carbon of the deoxyribose sugar which is required to bind a subsequent nucleotide. | |
Deoxyribonucleic Acid, a double stranded, helical molecule. | ||
DNA ladder | Mixture of DNA molecules of known base pair length. These provide a measure of how far a DNA molecule travels during gel electrophoresis. | |
Master mix | A mixture containing the common components for several PCRs, this is made in a large batch or master mix which is then divided between individual reactions. Master mixes contain enough reagents for the required number of tests, typically plus one to allow for pipetting errors. | |
Mix by pipetting | Drawing up and expelling a substance up to ten times using an automatic pipette, with the aim of mixing the solutions. | |
Water MB | Deionised, filtered and autoclaved water. | |
Negative control | A reaction which comprises all but one essential component, thereby proving the necessity of the absent substance. | |
PCR | Polymerase Chain Reaction - an enzyme driven reaction where DNA molecules are replicated. | |
Phred score | The likelihood that a base call in a DNA sequence is incorrect, a score of 20 has a 1 in 100 probability of being an incorrect call, 30 is 1 in 1000 etc. | |
Positive control | A reaction comprising all common PCR components and a known DNA sample, thereby proving the suitability of all reagents. | |
Primer (oligonucleotide) | ||
Probe hybridisation | The complementary binding of an oligonucleotide to a target DNA molecule causing a measurable response. | |
Sanger sequencing | The method by which a DNA sequence is resolved, developed by Frederick Sanger and colleagues. | |
Sequencing | Identifying the order of the nucleotide sequence of DNA. | |
Thermal cycler | The machine that performs the cycling of temperatures required for a PCR. | |